Leaving Community
Are you sure you want to leave this community? Leaving the community will revoke any permissions you have been granted in this community.
CELPP: Continuous Evaluation of Ligand Protein Prediction
Weekly blinded challenges for ligand pose prediction
|
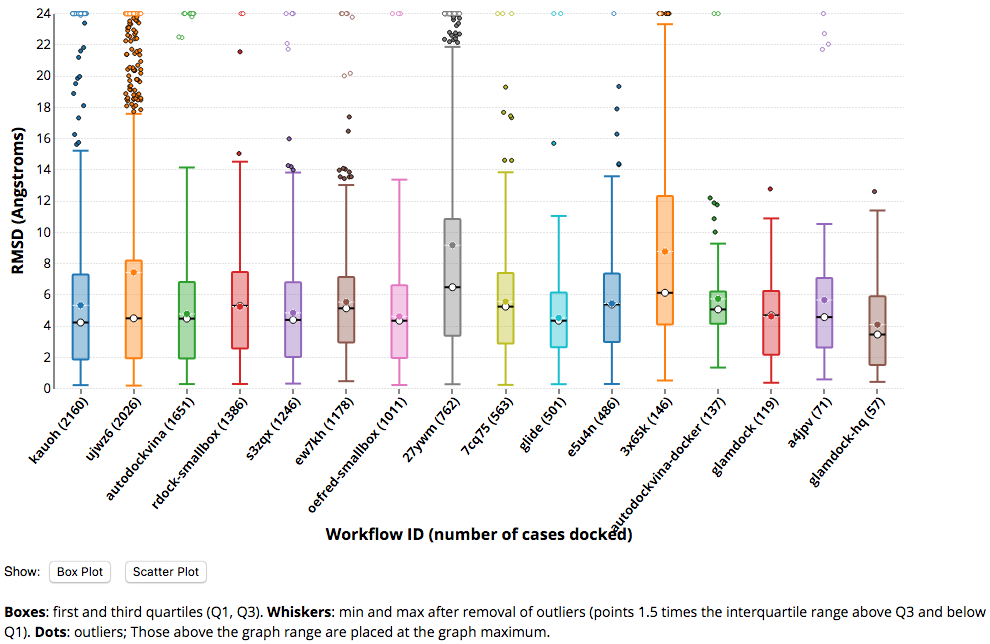
CELPP uses the Protein Data Bank's (PDB) weekly publication of a list of structures slated for imminent release into the public domain as the basis for a weekly, pose-prediction challenge. Participants in CELPP create a workflow to predict protein-ligand binding poses, which is then tasked with predicting 10-100 new (never before released) protein-ligand crystal structures each week. CELPP evaluates the accuracy of each workflow's predictions and posts the scores online.
For more details, please see our CELPP wiki, CELPP GitHub pages, or join our Google Group.
Using or publishing any CELPP data means that you agree to cite the following reference: Wagner, J., Churas, C., Liu, S., Swift, R., Chiu, M., Shao, C., Feher, V., Burley, S., Gilson, M., and R. Amaro, "Continuous Evaluation of Ligand Protein Predictions: A Weekly Community Challenge for Drug Docking" (2018) BioRxiv Preprint, DOI:10.1101/469940