Leaving Community
Are you sure you want to leave this community? Leaving the community will revoke any permissions you have been granted in this community.
Overview
Please note that a number of procedures have been changed, relative to prior challenges, to facilitate the processing and evaluation of predictions.
For Stage 2, you can submit predictions for protein-ligand affinity scores or rankings, and/or binding free energies for the small free energy (FE) challenge sets. Each prediction must be submitted in the form of a gzipped tar (.tgz) file. The D3R website will provide separate upload options for files of the following four types:
- A structure-based Score tgz file, containing ligand scores and/or ranks based on a docking/scoring method, and associated information.
- A ligand-based Score tgz file, containing ligand scores and/or ranks generated by a purely ligand-based prediction method, such as QSAR, and associated information.
- A Set 1 Free Energy file, containing absolute or relative free energy predictions for the first small FE compound set in the challenge, and associated information.
- A Set 2 Free Energy file, containing absolute or relative free energy predictions for the second small FE compound set in the challenge, and associated information.
Participants will select one of the above options with radio buttons on the D3R website.
If you make multiple predictions by different methods, such as by docking with several different energy functions, each prediction set must be in its own separate .tgz file.
For structure-based affinity and free energy predictions, the poses used must be provided along with the scores, even if the poses used for scoring are the same as the Stage 1 released structures. This is to simplify our recording and tracking of predictions. Although the poses in the Score tgz will not be evaluated, we ask that you provide them because they may be useful in explaining the quality of the scoring predictions.
SMILES string for ligand FXR_33: You may have noticed that the SMILES string for ligand FXR_33 does not match the ligand in the corresponding crystal structure. Roche scientists have explained to us that the ligand used to grow the crystal was in fact the N-oxide, but the pyridine appeared in the crystal structure instead. They attribute this either to oxidation occurring during the several days required for the crystal to grow, or possibly to the pyridine being present as an impurity (1% or less). We are considering whether to evaluate scoring of this ligand.
Also new in GC2, the names of the Score and Free Energy tgz files are now free-form; they are not required to include any particular text.
The following section details the required contents and format of these files. Additionally, blank template files for the two components of this challenge are available for download, as are examples of completed Score.tgz and Free_Energy.tgz files. Note that the example files contain artificial information, and thus serve only to illustrate the required contents and formats of a submission.
template files || structure based score example files || ligand based score example files || free energy set 1 example files || free energy set 2 example files
Submissions that do not adhere to these requirements should be rejected by our submission system; we recommend that you leave time before the deadline to correct any technical errors that result during the submission process. If a file with technical errors does pass the automated validation step, we will do our best to interpret the submission and may contact you for help with this. However, if a file proves particularly problematic, it may be necessary to omit the submission from evaluation.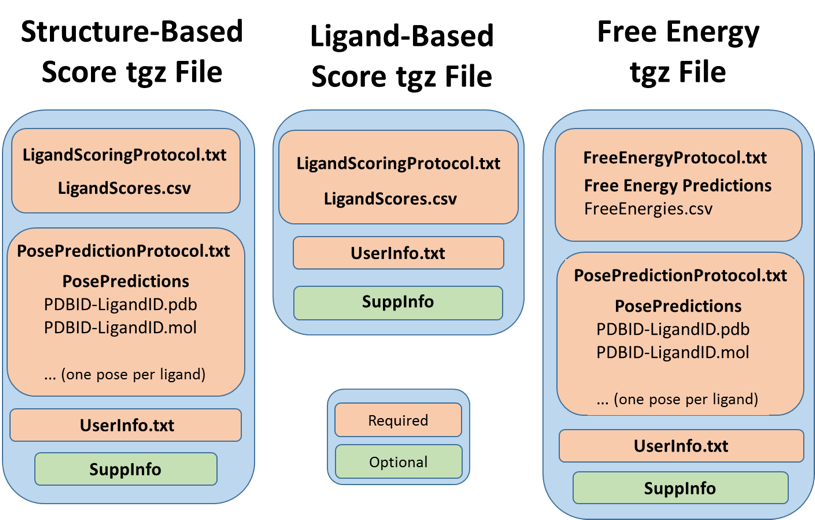
Figure 1. Required contents of the three types of tgz files for Stage 2 of Grand Challenge 2.
Content and Generation of TGZ files
The information required for each type of tgz file is summarized in Figure 1 and detailed in the following subsections. Note that every tgz type requires a user information file (Userinfo.txt). Because this file is common to all tgz types, it is not addressed in the descriptions of each tgz type, but is instead detailed separately (see The User Information File, below), as is the general procedure for generating the tar files (see How to make your Score and Free Energy tar files, below). In addition, we now encourage you to submit a Supplementary Information directory as part of each Score or Free Energy tgz file; this may contain any input, data, script or other files that would help to us interpret and/or reproduce your results. Potential contents of a Supplementary Information directory are listed in a subsection below.
A final subsection below touches on some of the common validation errors we have seen, and how to avoid them.
Structure-Based Score tgz File
A structure-based Score tgz file is used to submit one set of ligand scores or rankings generated by a structured-based method, along with information on how the predictions were made, the set of poses used, and the protocol used to generate the poses. A User Information file is required, and a Supplementary Information directory (see below) is optional but encouraged.
Each ligand scoring and ranking is described by two files: a ligand scoring protocol file, and a ligand scoring results comma-separated value (CSV) file, named LigandScoringProtocol.txt and LigandScore.csv respectively. The results file contains the scores and corresponding rankings generated by the protocol.
The ligand scoring protocol file must contain a brief, structured summary, in the form of a plain-text document, of how you scored the ligands according to predicted affinity for the target protein. A template file is provided for your convenience, as is a completed example file. Lines beginning with a hash-tag (#) may be included as comments. The file must contain the following components, as illustrated in the template and example:
- Your informal brief name for the protocol
- A list of the major software packages and their versions used in the protocol
- A listing of the key parameters used in the calculations
- A brief narrative of the procedure.
Each item must begin with the appropriate keyword; respectively:
- Name:
- Software:
- Parameters:
- Method:
The ligand scoring results file lists your rankings and scores or energies of the binding strengths of the ligands to the target protein. Again, lines beginning with # will be treated as comments. As some scoring methods provide results interpretable as binding energies or free energies, while others provide scores without well-defined units, the first non-comment line of your file must specify whether you are providing energies or scores. This line must take one of the following forms:
Type: energy
or
Type: score
If your results are given as energies, the units must be in units of kcal/mol.
Each subsequent non-comment line of the file comprises the identifier of one ligand; your ranking of the ligand within the set, where 1 corresponds to maximal affinity; and your computed binding energy, free energy, or score. These three items should be separated by commas.
The scoring file must contain a line for every ligand in the challenge. If you have not entered a prediction for a ligand, the corresponding line should have a placeholder: "inact" for compounds you identified as inactive, or "nopred" if you are not supplying a prediction for the compound for any reason.
Please refer to the template and example files; note that the template files are prefilled with the list of ligand identifiers, for your convenience.
The pose prediction protocol and the predicted poses should be provided, where only one pose is permitted for each protein-ligand pair, and this submitted pose should be the pose used to generate the score or ranking of the ligand. If you used multiple poses to generate ligand scores, please submit only one pose per ligand in this tgz file, and provide the rest of the poses in the Supplementary Information Directory (below).
The protocol file is named PosePredictionProtocol.txt, and it contains a brief, structured summary, in the form of a plain-text document, of your pose predictions methods. Lines beginning with a hash-tag (#) may be included as comments. The file must contain the following components, as illustrated in the template and example:
- Your informal, brief name for the protocol
- A list of the major software packages and their versions used for system preparation and pose prediction
- List of key parameters used for system preparation
- Plain-text description of system preparation method
- List of key parameters used for pose prediction
- Plain-text description of pose prediction method
Each item must begin with the appropriate keyword; respectively:
- Name:
- Software:
- System Preparation Parameters:
- System Preparation Method:
- Pose Prediction Parameters:
- Pose Prediction Method:
Each pose prediction must be provided in the form of a protein structure PDB file and a corresponding ligand V2000 MDL mol file (see, e.g., http://en.wikipedia.org/wiki/Chemical_table_file) with 3D atomic coordinates for the pose, where the coordinates in the protein PDB file and the ligand molfile are in the same frame of reference. Any ligand coordinates provided in PDB format or included in the protein PDB files will be ignored. You may treat the protein as rigid or flexible, but you must rotationally and translationally superimpose all of your final structure predictions onto the reference protein structure provided in the challenge data package in order to facilitate evaluation of your predictions. We request molfiles to prevent problems with the parsing of ligand coordinates in PDB format, which arose in Grand Challenge 2015.
The file names of your pose prediction protein PDB and ligand mol files must be constructed as follows:
<PDB ID of initial protein structure>-<LigandID>.pdb
<PDB ID of initial protein structure>-<LigandID>.mol
Here <PDB ID of initial protein structure> is the PDB ID of the structure that you docked the ligand into; for example, it might be 3OMM. LigandID is the identifier of the ligand for this challenge; for example, it might be FXR_1. Thus, your pose prediction for ligand FXR_1, generated by docking into structure 3OMM, would be contained in the following two files:
3OMM-FXR_1.mol
3OMM-FXR_1.pdb
If you used a Stage 1 crystal structure, such as 1HGAF.pdb, please leave the initial "1" off when constructing these filenames; e.g.,
HGAF-FXR_1.mol
HGAF-FXR_2.pdb
Sample pose prediction protein PDB files and ligand mol files are included with these instructions, as are example and template files for PosePredictionProtocol.txt.
Ligand-Based Score tgz File
A ligand-based Score tgz file is used to submit one set of ligand scores or rankings generated by a method that does not use ligand-protein poses, along with information on how the predictions were made. It has the same contents as a structure-based Score file (above), except that it does not include the poses and pose-prediction protocol files.
Set 1 and Set 2 Free Energy tgz Files
A Free Energy tgz file is used to submit predictions of absolute or relative binding free energies for one of the free energy sets in the challenge. If you ran calculations for both free energy sets, you should still submit them separately. Note that you are free to submit both ligand scores for the full set of compounds (above), and free energy calculations for these two smaller sets. A Free Energy tgz file should contain free energy predictions in a CSV file, FreeEnergies.csv; and a protocol file, FreeEnergyProtocol.txt, explaining the methodology. If your calculations involved pose predictions by a docking method, then the free energy file should also include these pose predictions. A User Information file is required, and a Supplementary Information directory (see below) is optional but encouraged.
The FreeEnergyProtocol file must contain a brief, structured summary, in the form of a plain-text document, of how you scored the ligands according to predicted affinity for the target protein. A template file is provided for your convenience, as is a sample filled-out file. Lines beginning with a hash-tag (#) may be included as comments. The file must contain the following components, as illustrated in the template and example.
- Your informal brief name for the protocol
- A list of the major software packages and their versions used in the protocol
- A listing of the key parameters used in the calculations
- A brief narrative of the procedure.
The FreeEnergies.csv file follows a format similar to that used for ligand scoring. Each line should list one ligand, followed by the predicted binding free energy (kcal/mol), and then by your estimate of the numerical uncertainty (standard error of the mean) in the prediction due to limitations in sampling. If you computed relative binding free energies, the results should all be referenced to the first ligand listed (see template), which hence should be assigned a free energy of 0.0 with zero uncertainty.
If your calculations relied on predicted poses, please provide these, along with the protocol used, as specified for the structure-based Score tgz file (above). Note that only one pose is permitted for each protein-ligand pair.
The User Information File
As noted above, every tgz file must include a User Information file. This is a text file named UserInfo.txt and containing five lines of text, as follows:
- Submitter Last Name:
- Submitter First Name:
- Submitter Email:
- Submitter Organization:
- Research Group or PI Name:
- Research Group or PI Email:
We are asking for this file to maximize clarity regarding the associations between submissions and submitters and research groups.
The Supplementary Information Directory
Every tgz file can optionally contain a Supplementary Information directory (folder), called SuppInfo, containing added files that would help interpret and reproduce your results. Examples of files you might provide include:
- Plain-text scripts used to drive the calculations
- Input parameters for the programs
- Intermediate results
- Output files (Files over 10MBs may be problematic, please contact us in case you're experiencing problems.)
The files may have any names you like. To include this directory in your .tgz file, just include a SuppInfo directory with your files within the directory you tar up.
How to Make Your Pose, Score, and Free Energy tgz Files
In order to enable automated processing of all submissions, we ask that you generate your Pose, Score and Free Energy tar files as follows:
- Put all files, and optionally your SuppInfo directory, into one directory having any name you like.
- Use tar options that prevent incorporation of the full pathname. (For example, the directory in your tar file should look like "/home/username/FXR/Pose".) The following is an example tar command which will not include this pathname information: tar -cvzf myPose.tar.gz --directory=/home/username/FXR Pose
- There should be no extraneous files, such as .sh scripts, Excel files, other tar files, etc, other than in the SuppInfo directory.
- For Mac users, be sure your tar file includes no extraneous Mac-specific files. For example, these might show up as "._ 3OMM-FXR_1-1.pdb" or ".DS_Store". The following example, tar command will generate a tar file without these extras: tar --disable-copyfile --exclude=.DS_Store -cvzf myPose.tar.gz Dock
Avoiding Common Errors
- The widely used program OpenBabel has a bad habit of changing the element Br and Cl to B and C in the molfiles it generates. If you use OpenBabel, please check for and correct these errors.
- Use a text editor to take a look at your molfiles before finalizing your submission. This should help you catch obvious problems, like molfiles with no atoms and molfiles with thousands of atoms, which do crop up occasionally.
- In the PosePredictionProtocol.txt, UserInfo.txt, LigandScoringProtocol.txt, FreeEnergyProtocol.txt files, please refer to the provided template files to ensure that you have included all the required lines and have not altered the field headings.
- If you are getting an error from using a filename with a 5 character PDB code, please just delete the "1" from the 5-letter codes to leave a 4-letter code. For example, 1hgaf--> hgaf.

