Leaving Community
Are you sure you want to leave this community? Leaving the community will revoke any permissions you have been granted in this community.
File Formats for Submitting Grand Challenge 2015 Predictions
October 24, 2015
For each challenge, you can submit the following types of predictions: ligand-protein poses, ligand affinity scores or rankings for the full set of ligands, and/or binding free energy calculations for the designated free energy subsets of the ligands. All predictions must be submitted in the form of gzipped tar (.tgz) files of three possible types:
- A dock file contains pose predictions and, optionally, structure-based ligand scores or ranks computed using these poses.
- A score file contains ligand scores or ranks, without any pose predictions; this is to allow for ligand-based prediction methods, such as QSAR.
- A free_energy file contains ligand binding affinities computed for the small “FEP” compound sets in the challenge. If your calculations for these sets involved pose predictions by a docking method, then the free energy file should include information on the pose predictions.
These files are summarized in the figure, and the subsequent text details their contents and format. Additionally, you can download completed example Dock.tgz, Score.tgz and Free_Energy.tgz files, as well as blank template files for pose prediction protocols, ligand scores, ligand scoring protocols, free energy protocols, and free energy predictions. The examples and templates are all based on the HSP90 challenge.
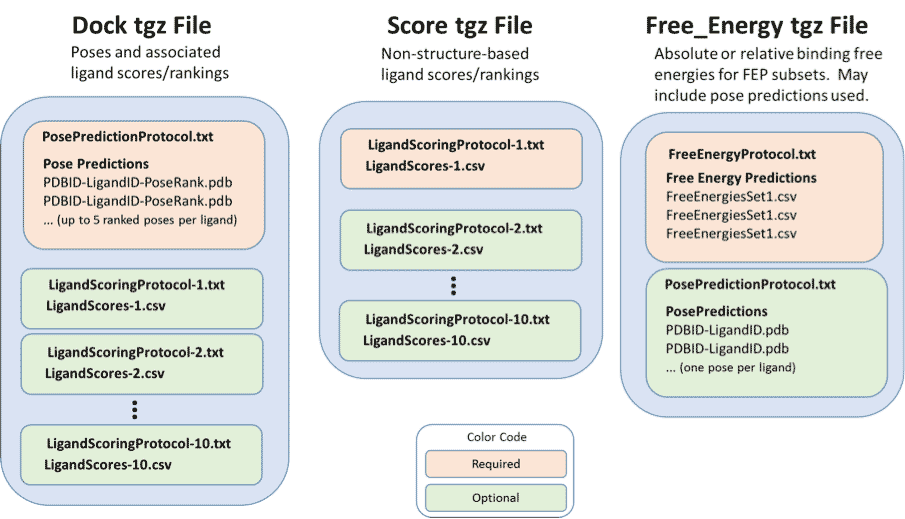
Figure 1. Diagrammed contents of the three types of tgz files for Grand Challenge 2015. See text for details.
Dock tgz file
The name of a dock/score tgz file must include the string “dock”, so that the file name has the form *dock*.tgz, except that the strings “free_energy” and “score” may not be in the file name (see below). The file must contain a description of and the results from one and only one pose-prediction protocol. Optionally, the file may also contain the protocols for and results from up to ten different ligand affinity scoring or ranking calculations, where the scores or ranks must be based on the poses in the same dock tgz file. If more than one scoring protocol and results set is included in the file, the files describing them must have distinct names. The following subsections detail the files contained in a dock .tgz file.
Pose prediction protocol and result files
One dock tgz file contains a single protocol file, and PDB files with up to 5 poses predicted by this protocol for each ligand. The dock pose prediction protocol file, named PosePredictionProtocol.txt, containing a brief, structured summary, in the form of a plain-text document, of how you did the pose predictions. Lines beginning with a hash-tag (#) may be included as comments. The file must contain the following components, as illustrated in the template and example.
- Your informal brief name for the protocol
- A list of the major software packages and their versions used in the protocol
- A listing of the key parameters used in the calculations
- A brief narrative of the procedure.
Each pose prediction must be provided in the form of a PDB file, containing both the entire protein and the entire ligand. Note that you may use any PDB file as your starting point, and you may treat the protein as rigid or flexible. However, you must rotationally and translationally superpose your final structure prediction onto one of the structures provided for this challenge, to facilitate evaluation of your predictions against the blinded crystal structures.
The file names of your pose prediction PDB files must be constructed as follows:
<PDB ID of protein structure>-<LigandID>-<poseRank>.pdb
Here <PDB ID of initial protein structure> is the PDB ID of the structure you docked the ligand into; for example, for HSP90, it might be 1YER. LigandID is the identifier of the ligand for this challenge; for example, it might be HSP90_1. And poseRank is the rank of this pose among the poses you predicted for this ligand, where 1 is best and 5 is worst; if you predicted only one pose, assign it a poseRank of 1. Thus, your second-ranked pose prediction for ligand HSP90_1, generated by docking into structure 1YER, would be
1YER-HSP90_1-2.pdb.
Each PDB pose prediction file must meet the following requirements:
- Each line with protein atom coordinates must start with ATOM
- Each line with ligand atom coordinates must start with HETATM
- All ligand heavy atoms must be present
- The ligand lines must be separated from the protein lines with a “TER” record
- The atom names assigned to the ligand atoms must start with the appropriate element symbols. For example, a nitrogen may not be called C25, but it may be called N10.
Additionally, if you submit multiple poses for a ligand, each prediction must include a remark line with either an energy (kcal/mol) or a score (arbitrary units), in the format
REMARK <energy/score> <value>
For example, this line might be
REMARK energy -20.6
REMARK score 5.7
Sample pose prediction PDB files are included with these instructions, as are example and template files for PosePredictionProtocol.txt.
Ligand scoring protocol and result files
As noted above, a given pose prediction protocol may be packaged in the same tgz file as up to ten different scorings or rankings based on the predicted poses. Each ligand scoring or ranking is described by two files: a ligand scoring protocol file, named LigandScoringProtocol-n.txt, where n is n integer from 1 to a maximum of 10; and a ligand scoring results file, named LigandScores-n.csv, which contains the scores and/or rankings generated by the corresponding protocol. These files are now described.
The ligand scoring protocol file must contain a brief, structured summary, in the form of a plain-text document, of how you scored the ligands according to predicted affinity for the target protein. A template file is provided for your convenience, as is a sample filled-out file. Lines beginning with a hash-tag (#) may be included as comments. The file must contain the following components, as illustrated in the template and example. The required components are the same as those for the pose prediction file, though the methodology used may well be different.
- Your informal brief name for the protocol
- A list of the major software packages and their versions used in the protocol
- A listing of the key parameters used in the calculations
- A brief narrative of the procedure.
The ligand scoring results file lists your rankings and scores or energies of the binding strengths of the ligands. Please refer to the template and example files. Again, lines beginning with # will be treated as comments.
Since some docking methods provide results interpretable as binding energies or free energies, while others provide scores without well-defined units, the first non-comment line of your file must state whether you are providing energies or scores. This line must take one of the following forms:
Type: energy
or
Type: score
If your results are given as energies, the units must be in units of kcal/mol.
Each subsequent non-comment line of the file comprises the identifier of one ligand for the protein target in question; your ranking of the ligand within the set, where 1 corresponds to maximal affinity; and your computed binding energy, free energy, or docking score. These three items should be separated by commas.
The template file is prefilled with the list of ligand identifiers, for your convenience.
Separate files will be used for the smaller “FEP” prediction sets (see below). However, if you used a free energy method for all of the compounds, you can use the present file format to document these calculations.
Score tgz file
A score tgz file is used to submit ligand scores or rankings that are not based on docking calculations and hence need not be associated with predicted poses. For example, you might train a QSAR method or a neural network using publicly available data on HSP90 inhibitors, and use the trained model to predict the activities of the challenge compounds. The name of a score tgz file must include the string “score”, so that the file name has the form *score*.tgz, except that the strings “free_energy” and “dock” may not be in the file name. The file contains the description of and results from one to ten ligand-scoring or ranking methods that are not dependent on computed poses. If more than one scoring protocol and results set is included in the file, the files describing them must have distinct names.
Each ligand scoring or ranking is described by two files: a ligand scoring protocol file, named LigandScoringProtocol-n.txt, where n is n integer from 1 to a maximum of 10; and a ligand scoring results file, named LigandScores-n.csv, which contains the scores and/or rankings generated by the corresponding protocol. Please create these according to the instructions given above in “Ligand scoring protocol and result files”.
Free_energy tgz file
If you used a free energy method, such as FEP or TI, to compute the absolute or relative binding free energies of the compounds in the small “FEP” compound sets, please submit these predictions in a free energy tgz file. The name of a free energy tgz file must include the string “free_energy”, so that the file name has the form *free_energy*.tgz, except that the strings “score” and “dock” may not be in the file name. Note that you are free to submit both ligand scores for the full set of compounds (above), and also to submit free energy calculations for any or all of thsee smaller sets. Each free_energy tgz file should include the results from a single free energy protocol for all of the compound sets that you applied this protocol.
The free energy calculation protocol file must be named FreeEnergyProtocol.txt, and it must contain a brief, structured summary, in the form of a plain-text document, of how you scored the ligands according to predicted affinity for the target protein. A template file is provided for your convenience, as is a sample filled-out file. Lines beginning with a hash-tag (#) may be included as comments. The file must contain the following components, as illustrated in the template and example. The required components are the same as those for the pose prediction file, though the methodology used may well be different.
- Your informal brief name for the protocol
- A list of the major software packages and their versions used in the protocol
- A listing of the key parameters used in the calculations
- A brief narrative of the procedure.
As shown in the free energy template and example files, the free energies for each set of compounds should be submitted in a format similar to that used for ligand scoring, and named according to set; e.g., FreeEnergiesSet1.csv, FreeEnergiesSet2.csv, FreeEnergiesSet3.csv. Thus, each line should list one ligand, followed by the predicted binding free energy (kcal/mol), and then by your estimate of the numerical uncertainty (standard error of the mean) in the prediction due to limitations in sampling. If you computed relative binding free energies, the results should all be referenced to the first ligand listed (see template), which hence should be assigned a free energy of 0.0 with zero uncertainty.
If these calculations are based on a set of computational pose predictions, then the file must also include the pose prediction protocol and pose predictions that were used, according to the instructions in the dock tgz file section above; and all of the free energy calculations in the file must correspond to this set of pose predictions. If you did some free energy calculations based on a different set of pose predictions, create a separate free energy .tgz file for those, with its own distinct pose prediction protocol and pose predictions.

