Leaving Community
Are you sure you want to leave this community? Leaving the community will revoke any permissions you have been granted in this community.
MAP4K4 - Overview
Challenge timeframe: Sep 15, 2015 to Feb 02, 2016
Welcome to D3R's inaugural Grand Challenge!
The Grand Challenges provide blinded unpublished datasets containing high quality crystal structures and binding affinity or potency data for testing and improving ligand-protein docking algorithms and their scoring protocols.
Each dataset is curated and embellished to challenge the methods, and their users, in a particular aspect of known docking protocol shortcomings.
Dataset 1
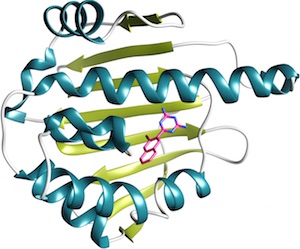
The HSP90 dataset, kindly donated by Abbvie, was expanded and curated by the CSAR group at University of Michigan. The ATP site of HSP90 has been the subject of many oncology drug discovery programs over the past 15 years and consequently has a large representation in the PDB and literature. However, the prevalence of water-mediated ligand protein-interactions and a ligand binding site that accesses multiple open and closed pocket conformations can make estimation of docking pose and ranked affinity a challenge.
This dataset has 8 crystal structures with resolution < 2.0Å, binding data for 180 compounds across five orders of magnitude and three chemical series, and over 50 inactives. The challenge has two stages, as follows:
Stage 1
Challenge: Predict the crystallographic poses of 6 ligands spanning all three chemical series, and predict affinities, or affinity rankings, for these ligands, and also for the other 174 ligands.
Provided Inputs: A) Protein-ligand co-crystal structures, 4 drawn from the PDB and 2 from the blinded dataset, that were solved with compounds of each chemical class in the series and prepared with hydrogens added. B) SMILES strings of the 6 ligands to be docked, of the 2 ligands in the newly revealed co-crystal structures, and of the additional 172 compounds for affinity prediction or ranking, C) benchmark IC50 values for relevant input structures. Note: There are Abbott publications for two of the chemical series of this Challenge containing crystal structures, SAR and related IC50s.
Outputs: A) Your predicted poses for the 6 ligands, in a coordinate system aligned with those provided in the Inputs. B) Your predicted affinities, or affinity rankings, for all 180 compounds.When Stage 1 closes, we will release the crystallographic poses of the 6 ligands.
Stage 2
Challenge: Predict the affinities, or affinity rankings, of all 180 ligands.
Inputs: Same as for Stage 1, supplemented by the co-crystal structures. Correction: IC50s will be released once Stage 2 is over.
Outputs: Your predictions of the affinities, or affinity rankings of all 180 compounds.N.B. You are free to use additional public-domain protein structures and scientific literature to help you make your predictions in both stages. For example, if you prefer to dock the ligands into a different structure from the PDB, this is fine, so long as the structures you submit as your predictions are rotated and translated so they superimpose on the structures we provided.
Dataset 2
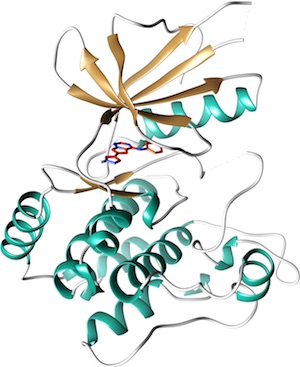
The MAP4K4 dataset, kindly donated by Genentech, has been curated by D3R.
This kinase, a member of the STE20 family, is involved in a number of pathways that are important to drug discovery, metabolism, cancer and inflammation.
This dataset comprises 18 compounds and crystal structures with resolution <2.5 Å and inhibition/binding data over four orders of magnitude, in many cases confirmed by multiple assay methods. There are one chemical series and a number of diverse compounds ranging in size from that of fragments with high ligand efficiency to drug-like, with 65% of the dataset having double digit nM inhibition. Fragment binding is confirmed by both SPR and crystallography. Conformational flexibility of the kinase P-loop will provide additional challenges to typical binding mode tests.
Similar to HSP90, the challenge has two stages:
Stage 1
Challenge: Predict the poses of 30 co-crystal structures containing diverse chemical series and affinity predictions/rankings of 18 of the ligand structures. Note: We have more co-crystal structures than affinity data for this target.
Provided inputs: A) Two protein-ligand co-crystal structures drawn from the PDB that represent the flexible nature of the target?s active site region. B) SMILES strings of the 30 ligands to be docked for pose prediction. The 18 ligands whose predictions/ranking is to be predicted are specified in the "Instructions and Descriptions" pdf document provided in the data package.
Output: Your predicted poses for ALL the 30 ligands in a coordinate system aligned with those provided in the Inputs. B) Your predicted affinities/rankings for the 18 compounds for which affinity data are available.
Stage 2
Challenge: Repeat the affinity predictions/rankings for the 18 ligands for which affinity data are available, now considering the disclosed ligand poses.
Inputs: The co-crystal structures of the aforementioned 18 ligands will be provided.
Outputs: Your predictions of the affinity predictions/rankings of the 18 ligands.
N.B. As with the case in the HSP90 target, you're free to use any public domain protein structures as long as the structures you submit as your predictions are rotated and translated so they superimpose on the structures we provided.
MAP4K4 - Data Download
Challenge timeframe: Sep 15, 2015 to Feb 02, 2016
MAP4K4 - Submissions
Challenge timeframe: Sep 15, 2015 to Feb 02, 2016
Please join the challenge and Login.
MAP4K4 - Evaluation Results
Challenge timeframe: Sep 15, 2015 to Feb 02, 2016
Evaluation Results
HSP90 |
MAP4K4 |
|
|---|---|---|
Spreadsheets (.csv)
Stage 1
Stage 2
Graphs: Stage 1 and 2 |
Spreadsheets (.csv)Graphs: Stage 1 and 2 |
Pose predictions were evaluated in terms of symmetry-corrected root-mean-square deviations (RMSD, Å). In the graphs, the pull-down menu allows display of the results for all submissions for each compound (e.g., MAP_01), or results for all submissions averaged over all compounds (AVG). The three buttons following the menu in the graphs allow display of the RMSD values for lowest RMSD pose among the maximum of five in each submission (Best); of the mean RMSD value across all five; or of the RMSD value for the top-scoring pose in each submission (Pose 1). The csv files provide the statistics averaged over ligands. Note that HSP90 compound 44 is excluded from pose prediction evaluations because of crystal artifacts found in the co-crystal structure. The rankings of compounds by affinity and the free energy calculations were evaluated in terms of the Kendall's tau and Spearman rho statistics, and the free energy calculations were additionally evaluated in terms of centered RMSDs (kcal/mol). Uncertainties in these statistics (e.g., Kendall Tau Err in csv files) were obtained by recomputing them in 10,000 rounds of resampling with replacement, where, in each sample, the experimental IC50 data were randomly modified based on the experimental uncertainties. The graphical representations provided here do not include Spearman's rho. Several participants submitted free energy values for all the compounds, rather than just the three small free energy datasets of HSP90 compounds. In those cases, the results are included in the "Score" evaluations. Further details of these procedures and results will be provided in an overview paper in the special issue of JCAMD. |

