Leaving Community
Are you sure you want to leave this community? Leaving the community will revoke any permissions you have been granted in this community.
Grand Challenge 4
Overview
Grand Challenge 4 (GC4) is a blinded prediction challenge for the computational chemistry community, with components addressing pose-prediction, affinity ranking, and free energy calculations. GC4 is based on two different protein targets, Cathepsin S (CatS) and beta secretase 1 (BACE). The datasets were generously contributed by Janssen Pharmaceuticals and Novartis, respectively.
The BACE component is associated with new cocrystal structures and hence includes pose-predictions, but there are no new cocrystal structures for CatS, so this component does not include pose-predictions. Both the CatS and BACE components include affinity ranking and free energy prediction challenges. The BACE free energy set involves scaffold hopping, while the CatS free energy set focuses on a single chemical scaffold. We expect that all compounds in both the BACE and CatS free energy sets had a charge of +2 at the assay pH values of 4.5 (BACE) and 5.0 (CatS).
Further information on each subchallenge is provided below, and the left-hand menu provides links to the data packages for each subchallenge. Note that you are free to use the scientific literature and any public-domain protein structures and affinity datasets to help with your predictions.
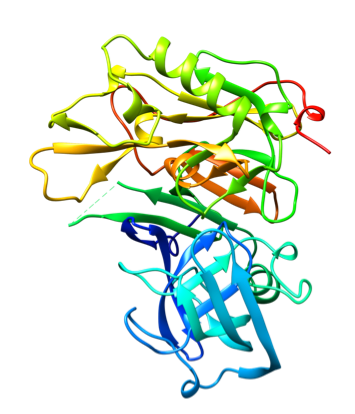
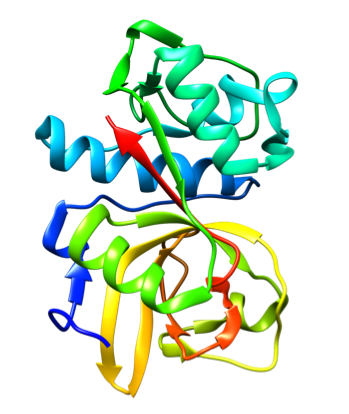
Representative crystal structures of BACE (PDB 5YGX) and CatS (PDB 2HHN).
Subchallenges
Subchallenge 1: BACE
This is a pose-prediction, affinity ranking, and free energy challenge, occurring in two stages. It is based on a dataset comprising 20 ligand-protein co-crystal structures, and binding data (IC50s) spanning three orders of magnitude for 154 compounds.
- Challenge:
Stage 1a: predict the crystallographic poses of 20 ligands. Predict affinities, or affinity rankings, for 154 ligands and/or the absolute or relative binding affinities for the designated free energy set of 34 compounds.
Stage 1b: predict the crystallographic poses of the 20 ligands in a self-docking challenge with the corresponding receptor structures. No affinity calculations in this stage.
Stage 2: predict affinities, or affinity rankings, for 154 ligands and/or the absolute or relative binding affinities for the free energy set of 34 compounds, this time taking advantage of the poses of the 20 compounds for which cocrystal structures will be released on October 22.
- Inputs:
Stage 1a: SMILES strings of the 20 ligands to be docked and the FASTA sequence of the target, BACE. SMILES strings of the 154 compounds for affinity prediction or ranking. SMILES strings of the molecules in the free energy set (34 molecules) for the calculation of relative or absolute binding affinities.
Stage 1b: SMILES strings of the 20 ligands to be docked, and the receptor structures cocrystallized with each ligand.
Stage 2: the same inputs as for Stage 1a, supplemented by the 20 cocrystal structures.
- Outputs:
In Stage 1a, your predicted poses for the 20 ligands, in a coordinate system of the participant's choosing rotationally and translationally superimposed onto the reference protein structure (5ygx, chain A). Your predicted affinities, or affinity rankings, for all 154 compounds and/or your predicted absolute or relative binding affinities (in kcal/mol) for the free energy set of 34 compounds.
In Stage 1b, predicted crystallographic poses of the 20 ligands. When Stage 1b closes, we will release the crystallographic poses of the 20 ligands.
In Stage 2, your predictions of the affinity rankings of all 154 compounds and/or absolute or relative binding affinities (in kcal/mol) for the free energy set of 34 compounds. In Stage 2, these calculations are performed with full knowledge of the cocrystal structures.
- Dataset name: BACE
- Subchallenge timeline:
- Stage 1a:
- September 4th- October 4th
- Receptor structures (without ligand) released on October 8th
- Stage 1b:
- October 8th - October 19th 17:00PST
- Cocrystallographic structures released on October 22nd
- Stage 2:
- October 22th - December 4th 17:00PST
Subchallenge 2: Cathepsin S
This is an affinity ranking, and free energy challenge. It is based on binding data (IC50s) spanning three orders of magnitude for 460 compounds.
- Challenge:
Predict affinities, or affinity rankings, for 459 ligands and/or the absolute or relative binding affinities for the designated free energy set of 39 molecules.
- Inputs:
FASTA sequence for the CatS target and the corresponding SMILEs strings of all 459 molecules for affinity prediction/ranking, SMILEs strings of the 39 molecules in the free energy challenge
- Outputs:
Predicted affinity/ranking for all 459 compounds, and the predicted affinities for all 39 molecules in the free energy set.
- Dataset names: CatS
- Subchallenge timeline: September 4th - December 4th

