Leaving Community
Are you sure you want to leave this community? Leaving the community will revoke any permissions you have been granted in this community.
PL-2016-1 - Overview
Challenge timeframe: Feb 15, 2016 to May 10, 2016
Updates
- 2016-04-11
- Submission instructions published
- 2016-04-11
- End date changed to May 09, 2016 17:00PST
- 2016-03-16
- End date changed to April 12, 2016
- 2016-03-02
- 3 diagrams added
- 2016-02-26
- Corrected datafile added
This challenge is based on two datasets, kindly donated by Barry Stoddard (Fred Hutchinson Cancer Research) and David Baker (U. of Washington), which center on proteins designed with the Rosetta program. One protein has been shown to bind 17-hydroxyprogesterone (17-OHP), and the other binds cholecalciferol (vitamin D3) and its analogue 25-hydroxycholecalciferol (25-D3).
The 17-OHP binding dataset contains two protein-ligand co-crystal structures. In both cases, the ligand is 17-OHP, and it is the designed proteins that differ. The two proteins share 93% sequence identity, and a good number of the sequence differences are in the binding site. The crystal structures, named PL-2016-1-O-1 and PL-2016-1-O-2, are solved at resolutions of 2.5 and 2.0Å respectively. Water-mediated protein-ligand interactions, involving a few conserved water molecules in the active site, are important in binding. Getting the conformations correct for the non-planar ring puckers in the 17-OHP ligand should add a layer of challenge for this dataset. These facts, coupled with the challenge of computing the difference between the affinities of the two proteins for their common ligand, 17-OHP, should make for an enlightening docking and scoring challenge.
The cholecalciferol binding dataset contains three protein-ligand co-crystal structures, each for a different designed protein variant bound to the ligand 25-D3. The three proteins have sequence identities > 90%, with subtle differences in the binding site. The crystal structures, named PL-2016-1-C-1, PL-2016-1-C-2 and PL-2016-1-C-3, have resolutions 1.9, 2.1 and 1.9 Å respectively. The corresponding binding affinities have been measured, and the affinity of the protein in PL-2016-1-C-1 for vitamin D3 is also available, albeit without a corresponding crystal structure. Water-mediated protein-ligand interactions within the active site are important for binding.
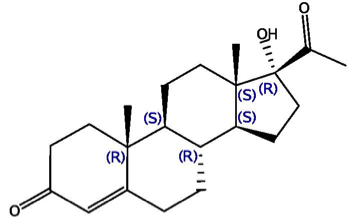 |
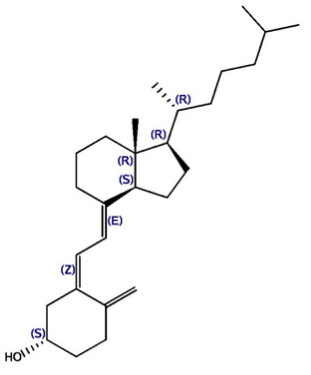 |
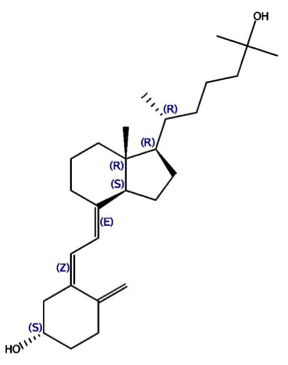 |
| 17-OHP | Vit D3 | 25-D3 |
Challenge:
Predict the poses of the 17-OHP and 25-D3 ligands with their respective proteins, as well as the affinity or affinity rankings for each dataset, including the affinity/ranking of the protein in PL-2016-1-C-1 with ligands 25-D3 and vitamin D3. Note that this challenge has only one stage.
Provided Inputs:
A) Protein structures with the co-crystalized ligand deleted and all the waters retained. No further preparation of the protein structures was done. B) SDfiles for ligands 17-OHP, D3 and 25-D3.
Outputs:
A) Your predicted poses, in a coordinate system aligned with those provided in the inputs, for the following complexes: PL-2016-1-O-1/17-OHP and PL-2016-1-O-2/17-OHP, PL-2016-1-C-1/25-D3, PL-2016-1-C-2/25-D3, and PL-2016-1-C-3/25-D3. B) Your predicted affinities, scores, or affinity rankings for the protein-ligand pairs in each of the two datasets; i.e., for all of the pose-prediction cases plus the affinity for PL-2016-1-C-2/vitamin D3. Each dataset may be ranked separately.
PL-2016-1 - Data Download
Challenge timeframe: Feb 15, 2016 to May 10, 2016
PL-2016-1 - Submissions
Challenge timeframe: Feb 15, 2016 to May 10, 2016
Please join the challenge and Login.
PL-2016-1 - Evaluation Results
Challenge timeframe: Feb 15, 2016 to May 10, 2016
Evaluation Results
The two tar files PL-2016-1-answers-corrections-7-25-16.tgz and PL-2016-1-answers.tar.gz provide a corrected set of answers for the PL-2016-1 challenge, and our evaluation of the submitted predictions.
The corrections involve swapping two affinities, and providing all four co-crystal structures from the asymmetric unit of one of the pose predictions, for which we previously provided only one. (See included readme file for details.) We are sorry for the initial mixup.
Evaluations are provided for both scoring and pose predictions, by receipt ID. The corrected results are also available on the PL-2016-1 web-page after login, and the evaluation files will also be posted there soon. The readme file with the evaluations explains how we handled pose evaluation for the case with four co-crystal structures in the asymmetric unit.

