Leaving Community
Are you sure you want to leave this community? Leaving the community will revoke any permissions you have been granted in this community.
D3R Grand Challenge 2
Grand Challenge 2
2017-07-14 11:00PST - New overview results are available
2017-03-28 14:00PST - Evaluation Results and user submitted files are available
2017-01-24 15:30PST - Stage 2 submission deadline has been extended to 2017-02-08 17:00PST
2017-01-18 16:30PST - Stage 2 submission system is live.
2017-01-10 15:00PST - Stage 2 Submission Instructions with template and example files are online
2016-11-22 17:00PST - Stage 1 submission window closed, Stage 2 data package with stage 1 poses available
2016-11-21 16:13PST - Submission window has been extended 24hrs to 2016-11-22 17:00PST
2016-11-18 9:49PST - upload size issue resolved.
2016-11-18 8:42PST - looking into upload size issues.
2016-11-18 8:30PST - PDB / ligand issue resolved
2016-11-18 8:00PST - submission code is not accepting more than one PDB per ligand, which is a bug. developer working on a fix now.
Are the peptides present in the binding assays?
The binding is from a scintillation proximity readout. There is no mention of the co-activator peptide in the assay protocol so it must be concluded that it was absent. The GST-fused FXR ligand binding domain was used in the SPA.
For crystallization purposes the co-activator peptide was used as a variable, and the structure was determined from whatever complex crystallized first:
ENALLRYLLDKD is the GRIP-1 peptide
DHQLLRYLLDKD is the NCOA-1 peptide
The two structures you cite are from isomorphous crystals, so either peptide should have worked. Alas, crystallization is so unpredictable.
1sjpr CRYST1 93.940 93.940 47.798 90.00 90.00 120.00 P 65
1kjyp CRYST1 93.641 93.641 48.144 90.00 90.00 120.00 P 65
The peptide itself is a mini-version of the transactivator protein (>1000 aa) and stabilizes the FXR-ligand complex
SMILES string for ligand FXR_033
You may have noticed that the SMILES string for ligand FXR_033 does not match the ligand in the corresponding crystal structure. Roche scientists have explained to us that the ligand used to grow the crystal was in fact the N-oxide, but the pyridine appeared in the crystal structure instead. They attribute this either to oxidation occuring during the several days required for the crystal to grow, or possibly to the pyridine being present as an impurity (1% or less). We will not include this ligand when evaluating your pose predictions, and we are considering whether to evaluate scoring of this ligand.
This Challenge provides a blinded unpublished dataset containing high quality crystal structures and potency data for testing and improving ligand-protein docking algorithms and their scoring protocols. It is based on the Farnesoid X receptor (FXR) target; the dataset is kindly contributed by Roche and curated by D3R.
The FXR nuclear receptor forms a heterodimer with RXR when activated, and binds to hormone response elements on DNA, leading to up- or down-regulation of the expression of certain genes. FXR agonists are regarded as potential therapeutics for dyslipidemia and diabetes.
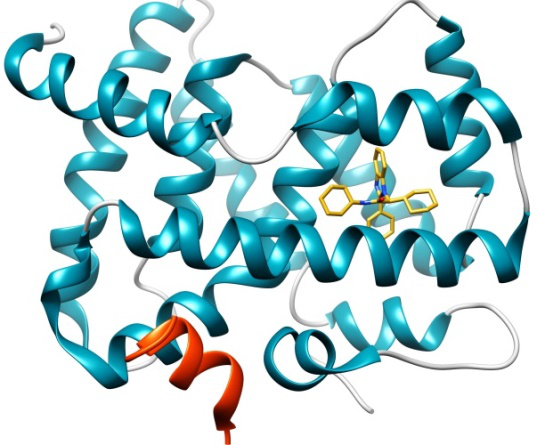
This dataset has 36 crystal structures with resolution < 2.6Å; and binding data (IC50s) for 102 compounds across five orders of magnitude (Scintillation Proximity Assay), comprising four chemical series and 6 miscellaneous compounds. The challenge has two stages, as follows:
Challenge - Stage 1
Challenge:
Predict the crystallographic poses of 36 ligands spanning all chemical series. Predict affinities, or affinity rankings, for these ligands, and also for the other 66 ligands. Predict the relative binding affinities for two designated free energy subsets of 18 and 15 compounds.Inputs:
The apoprotein as provided by Roche, with no further preparation of the protein structure done. This is not necessarily meant for use in your predictions, only for superimposing your pose prediction structures to facilitate evaluation. SMILES strings and SDfiles of the 36 ligands to be docked, and of the additional 66 compounds for affinity prediction or ranking. SMILES strings and SDfiles of two subsets (18 and 15 compounds) for the calculation of relative binding affinities by alchemical methods, such as free energy perturbation.Note: There are Roche publications for two of the chemical series of this Challenge containing crystal structures, SAR and related IC50s.1-3
Outputs:
Your predicted poses for the 36 ligands, in a coordinate system aligned with the apoprotein provided in the inputs. Your predicted affinities, or affinity rankings, for all 102 compounds. Your predicted relative binding affinities (in kcal/mol) for the free energy subsets of 18 and 15 compounds.When Stage 1 closes, we will release the crystallographic poses of the 36 ligands.

