Leaving Community
Are you sure you want to leave this community? Leaving the community will revoke any permissions you have been granted in this community.
FXR - Overview
Challenge timeframe: Sep 19, 2016 to Feb 08, 2017
This Challenge provides a blinded unpublished dataset containing high quality crystal structures and potency data for testing and improving ligand-protein docking algorithms and their scoring protocols. It is based on the Farnesoid X receptor (FXR) target; the dataset is kindly contributed by Roche and curated by D3R.
The FXR nuclear receptor forms a heterodimer with RXR when activated, and binds to hormone response elements on DNA, leading to up- or down-regulation of the expression of certain genes. FXR agonists are regarded as potential therapeutics for dyslipidemia and diabetes.
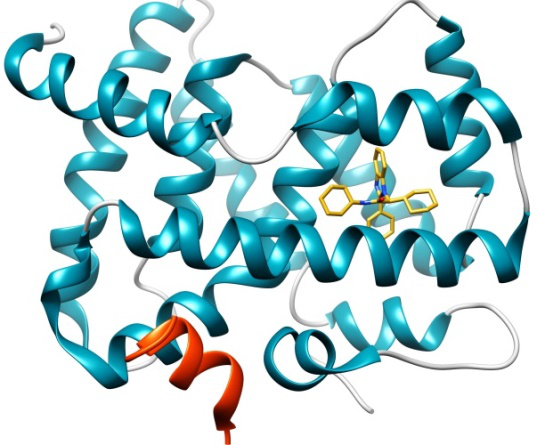
This dataset has 36 crystal structures with resolution < 2.6Å; and binding data (IC50s) for 102 compounds across five orders of magnitude (Scintillation Proximity Assay), comprising four chemical series and 6 miscellaneous compounds. The challenge has two stages, as follows:
Challenge - Stage 1
Challenge:
Predict the crystallographic poses of 36 ligands spanning all chemical series. Predict affinities, or affinity rankings, for these ligands, and also for the other 66 ligands. Predict the relative binding affinities for two designated free energy subsets of 18 and 15 compounds.Inputs:
The apoprotein as provided by Roche, with no further preparation of the protein structure done. This is not necessarily meant for use in your predictions, only for superimposing your pose prediction structures to facilitate evaluation. SMILES strings and SDfiles of the 36 ligands to be docked, and of the additional 66 compounds for affinity prediction or ranking. SMILES strings and SDfiles of two subsets (18 and 15 compounds) for the calculation of relative binding affinities by alchemical methods, such as free energy perturbation.Note: There are Roche publications for two of the chemical series of this Challenge containing crystal structures, SAR and related IC50s.1-3
Outputs:
Your predicted poses for the 36 ligands, in a coordinate system aligned with the apoprotein provided in the inputs. Your predicted affinities, or affinity rankings, for all 102 compounds. Your predicted relative binding affinities (in kcal/mol) for the free energy subsets of 18 and 15 compounds.When Stage 1 closes, we will release the crystallographic poses of the 36 ligands.
Challenge - Stage 2
Challenge:
Predict the affinities, or affinity rankings, of all 102 ligands and the relative binding affinities for the two free energy subsets of 18 and 15 compounds.Inputs:
Same as for Stage 1, supplemented by the 36 co-crystal structures. IC50s will be released once Stage 2 is over.Outputs:
Your predictions of the affinities, or affinity rankings of all 102 compounds as well as relative binding affinities (in kcal/mol) for the free energy subsets of 18 and 15 compounds, that is, if you chose to do or re-do the free energy predictions in this stage. N.B. You are free to use any public-domain protein structures and scientific literature to help you make your predictions in both stages, as long as the structures you submit as your predictions are rotated and translated so they superimpose on the apoprotein structure we provided.References
Richter, H. G.; Benson, G. M.; Bleicher, K. H.; Blum, D.; Chaput, E.; Clemann, N.; Feng, S.; Gardes, C.; Grether, U.; Hartman, P.; Kuhn, B.; Martin, R. E.; Plancher, J. M.; Rudolph, M. G.; Schuler, F.; Taylor, S. Bioorganic & medicinal chemistry letters 2011, 21, 1134. Richter, H. G.; Benson, G. M.; Blum, D.; Chaput, E.; Feng, S.; Gardes, C.; Grether, U.; Hartman, P.; Kuhn, B.; Martin, R. E.; Plancher, J. M.; Rudolph, M. G.; Schuler, F.; Taylor, S.; Bleicher, K. H. Bioorganic & medicinal chemistry letters 2011, 21, 191. Feng, S.; Yang, M.; Zhang, Z.; Wang, Z.; Hong, D.; Richter, H.; Benson, G. M.; Bleicher, K.; Grether, U.; Martin, R. E.; Plancher, J. M.; Kuhn, B.; Rudolph, M. G.; Chen, L. Bioorganic & medicinal chemistry letters 2009, 19, 2595.FXR - Data Download
Challenge timeframe: Sep 19, 2016 to Feb 08, 2017
FXR - Submissions
Challenge timeframe: Sep 19, 2016 to Feb 08, 2017
Please join the challenge and Login.
FXR - Evaluation Results
Challenge timeframe: Sep 19, 2016 to Feb 08, 2017
Evaluation Results
| Last updated Dec 20, 2017 | ||
|---|---|---|
OverviewsThe Overviews section is grouped by "prediction method".The previous spreadsheets and graphs, which were grouped by "submission type", can be found here
Stage 1
Pose Predictions (partials)
Stage 2
Scoring (partials) User Submitted Files
Stage 1
Stage 1 Pose Prediction submissions (355MB)
Stage 2
Stage 2 Scoring submissions (418MB) |
Evaluation Methods
Pose predictions were evaluated in terms of symmetry-corrected root-mean-square deviations (RMSD, Å). In the graphs, the pull-down menu allows display of the results for all submissions for each compound (e.g., FXR_1), or results for all submissions averaged over all compounds (AVG). The three buttons following the menu in the graphs allow display of the RMSD values for lowest RMSD pose among the maximum of five in each submission (Best); of the average RMSD value across all five (Average); or of the RMSD value for the top-scoring pose in each submission (Pose 1). The csv files provide the statistics averaged over ligands. Note that FXR_33 is excluded from pose prediction evaluations because of crystal artifacts found in the co-crystal structure. The rankings of compounds by affinity and the free energy calculations were evaluated in terms of the Kendall's tau and Spearman rho statistics, and the free energy calculations were additionally evaluated in terms of centered RMSEs (kcal/mol) and Pearson's r. Uncertainties in these statistics (e.g., Kendall Tau Err in csv files) were obtained by recomputing them in 10,000 rounds of resampling with replacement, where, in each sample, the experimental IC50 data were randomly modified based on the experimental uncertainties. The graphical representations provided here do not include Spearman's rho or Pearson's r. All free energy submissions were alchemical approaches that used explicit solvent free energy methods and provided relative binding free energies between pairs of ligands across the two challenge Stages and the two free energy sets, except for one submission, xk67c, which computed the absolute binding free energy of each separate ligand. In addition to the free energy methods, other approaches were applied to the free energy sets as part of the free energy component of the challenge; these included methods based on scoring functions, force fields combined with implicit solvent, and electronic structure calculations with implicit solvent. The two free energy ligand sets are part of the full series of 102 ligands, for which a variety of additional methods were tested in the affinity ranking component of the challenge. This meant that we could extract the predictions for the free energy sets of compounds from the larger set of ranking and scoring methods used for the 102 compound set, and thus put the more detailed free energy methods into the context of the faster methods that were applied to the full set of 102 ligands. Therefore, in the accompanied graphs and tables, RMSEc statistics are calculated for predictions from scoring methods with free energy estimates in the affinity ranking component of the challenge, and free energy methods in the free energy component of the challenge; and Kendall?s tau statistics are calculated for predictions from all scoring and free energy methods. Further details of these procedures and results will be provided in an overview paper in the special issue of JCAMD. All scripts used to evaluate submissions are publicly available on Github. Note regarding racemic compoundsThe instructions for this challenge stated that some of the compounds were tested by Roche as 50:50 racemates, and that there was evidence that the activity derived from only one of the enantiomers. For a 50:50 racemate, this would mean the raw IC50 is twice the IC50 of the active enantiomer. However, the instructions stated that participants should provide predictions that could be compared directly with the raw data. For these compounds, therefore, we have used your predictions without any adjustment to the raw experimental IC50 values. The compounds treated in this manner are as follows: FXR_37, FXR_39, FXR_40, FXR_42, FXR_50, FXR_51, FXR_52, FXR_53, FXR_54, FXR_55, FXR_56, FXR_57, FXR_58, FXR_59, FXR_60, FXR_61, FXR_62, FXR_63, FXR_64, FXR_66, FXR_67, FXR_68, FXR_69, FXR_70, FXR_71, FXR_72. By our accident, the instructions did not state that some other compounds also were tested experimentally as racemates. The following compounds were tested as 50:50 mixtures: FXR_6, FXR_7, FXR_8, FXR_9, FXR_13, FXR_18, FXR_20, FXR_22, FXR_25, FXR_31, FXR_35, FXR_36. For these compounds, we have divided the experimental IC50 values by 2 before comparing with your predictions. In addition, FXR_22 was tested as a 25:25:25:25 mixture of stereoisomers, so we have divided its experimental IC50 value by 4 before comparing with your predictions. Revision 2017-07-7 : Free energy evaluations were updated with ranking and scoring methods from the affinity ranking component of the challenge as detailed above, in order to put the more detailed free energy methods into the context of the faster methods that were applied to the full set of 102 ligands. The originally posted results are below. Originally posted resultsSpreadsheets (.csv)GraphsRevision 2017-04-20 : Error statistics were updated after we received further details regarding uncertainties in the experimental IC50 values, as well as a small correction. The originally posted results are below. Obsolete ResultsSpreadsheets (.csv)Graphs |

