Leaving Community
Are you sure you want to leave this community? Leaving the community will revoke any permissions you have been granted in this community.
BACE - Overview
Challenge timeframe: Sep 04, 2018 to Dec 04, 2018
Updates
2019-02-21 - Preliminary Affinity Ranking results are now available on the "Data Download" page. You must be logged in first to access the files.
2018-12-13 - Participant files are now available on the "Data Download" page. You must be logged in first to access the files.
2018-12-13 - Participant files are now available on the "Data Download" page. You must be logged in first to access the files.
2018-12-07 - Answers to the challenges are now available: BACE_score_compounds_D3R_GC4_answers.csv and BACE_FEset_compounds_D3R_GC4_answers.csv
2018-10-11 - An update was made to the Challenge submission instructions for Stage 1B. Also, for Stage 1B, when appropriate, please select your protocol from Stage 1A rather than reuploading a new protocol file.
Challenge submission instructions are available. Example and template files are also available.- BACE_target_D3R_GC4.fasta: Protein sequence file of the BACE construct used.
- BACE_pose_compounds_D3R_GC4.csv: CSV file of 20 compounds and their corresponding SMILES.
- BACE_score_compounds_D3R_GC4.csv: CSV file of 154 compounds and their corresponding SMILES string.
- BACE_FESet_compounds_D3R_GC4.csv: CSV file of 34 compounds for explicit-solvent relative or absolute free energy calculations.
- 5ygx, chain A: Reference protein structure to use for superimposition. Download: https://drugdesigndata.org/upload/community-components/5YGX_chainA.pdb. Note: The original PDB file has been altered to remove the 3 negative residues at the beginning and the residue numbered 0.
Introduction to BACE
Beta-Secretase 1 (BACE) is a transmembrane aspartic-acid protease human protein encoded by the BACE1 gene. BACE is essential for the generation of beta-amyloid peptide in neural tissue1, a component of amyloid plaques widely believed to be critical in the development of Alzheimer's, rendering BACE an attractive therapeutic target for this devastating disease2.
The BACE dataset provided herein comprises small molecule inhibitors across a three order of magnitude (nM to μM) range of IC50s along with previously undisclosed crystallographic structures. Specifically, we provide 154 BACE inhibitors for affinity, 20 for pose, and 34 for free energy prediction. This dataset was kindly provided by Novartis.
BACE Assay Conditions
A description of the assay conditions employed for GC4 BACE affinity data generation can be found in the following reference3.
BACE Crystallization Conditions
A detailed description of the crystallization conditions employed for GC4 BACE crystal structure generation can be found in the supplementary material of the following reference4.
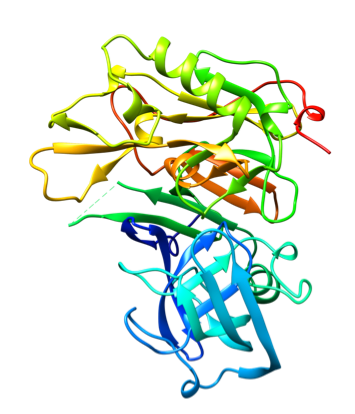
Representative crystal structures of BACE (PDB 5YGX).
References
- Vassar, R.; Kovacs, D. M.; Yan, R.; Wong, P. C. J. Neurosci. 2009, 29 (41), 12787-12794.
- Prati, F.; Bottegoni, G.; Bolognesi, M. L.; Cavalli, A. J. Med. Chem. 2018, 61 (3), 619-637.
- Hanessian, S.; Yang, G.; Rondeau, J.-M.; Neumann, U.; Betschart, C.; Tintelnot-Blomley, M. J. Med. Chem. 2006, 49 (15), 4544-4567.
- Machauer, R.; Laumen, K.; Veenstra, S.; Rondeau, J.-M.; Tintelnot-Blomley, M.; Betschart, C.; Jaton, A.-L.; Desrayaud, S.; Staufenbiel, M.; Rabe, S.; Paganetti, P.; Neumann, U. Bioorg. Med. Chem. Lett. 2009, 19 (5), 1366-1370.
BACE - Data Download
Challenge timeframe: Sep 04, 2018 to Dec 04, 2018
BACE - Protocols
Challenge timeframe: Sep 04, 2018 to Dec 04, 2018
Please join the challenge and Login.
BACE - Submissions
Challenge timeframe: Sep 04, 2018 to Dec 04, 2018
Please join the challenge and Login.
BACE - Evaluation Results
Challenge timeframe: Sep 04, 2018 to Dec 04, 2018
Evaluation Results
Overviews
Last updated April 18, 2019Pose Predictions
Affinity Predictions
This section presents metrics of the ability of the predictions to correctly rank ligands by affinity. The rankings were evaluated in terms of the Kendall's τ and Spearman's ρ. Predicted binding energies for the free energy sets were additionally evaluated in terms of centered root-mean-square error (RMSEc, kcal/mol) [1] and Pearson's R. Uncertainties in these statistics (e.g., Kendall's τ Errors in the table) were obtained by recomputing them in 10,000 rounds of resampling with replacement, where, in each sample, the experimental IC50 or Kd data were randomly modified based on the experimental uncertainties. Experimental uncertainties are added to the free energy, ΔG, as a random offset δG drawn from a Gaussian distribution of mean zero and standard deviation RTln(Ierr). In this evaluation, the value of Ierr was set to 2.5.
User Submissions
BACEStage1a.zip (812M)
BACEStage1b.zip (383M)
BACEStage2.zip (786M)
CatS.zip (966M)
gc4protocols.zip (700k)
Further details of these procedures and results will be provided in an overview paper in the special issue of JCAMD.
All scripts used to evaluate submissions are publicly available on Github.
(partials) indicates submissions that do not include the full set of predictions

